The mr_forest function draws a forest plot of causal estimates. The default option plots the variant-specific causal estimates (by/bx) and the estimate from the mr_ivw function using default settings (assuming variants are uncorrelated, random-effects for 4+ variants). Options allow users to plot estimates from a variety of different methods.
Usage
mr_forest(
object,
alpha = 0.05,
snp_estimates = TRUE,
methods = "ivw",
ordered = FALSE
)
# S4 method for MRInput
mr_forest(
object,
alpha = 0.05,
snp_estimates = TRUE,
methods = "ivw",
ordered = FALSE
)Arguments
- object
An
MRInputobject.- alpha
The significance level used to calculate the confidence intervals. The default value is 0.05, corresponding to 95% confidence intervals.
- snp_estimates
Whether to plot the variant-specific estimates. Defaults to
TRUE.- methods
Takes a string of computation methods used to calculate estimates. Defaults to
"ivw". Options are:"median"(simple median estimate),"wmedian"(weighted median estimate),"egger"(MR-Egger estimate),"mbe"(mode-based estimate),"conmix"(contamination mixture estimate), and"maxlik"(maximum likelihood estimate).- ordered
Determines by whether to arrange the variant-specific estimates in ascending order. Defaults to
FALSE.
Details
As the function produces a ggplot object, graphical parameters can be changed by adding commands from the ggplot2 package.
Examples
mr_forest(mr_input(bx = ldlc, bxse = ldlcse, by = chdlodds, byse = chdloddsse),
alpha = 0.01, ordered = TRUE)
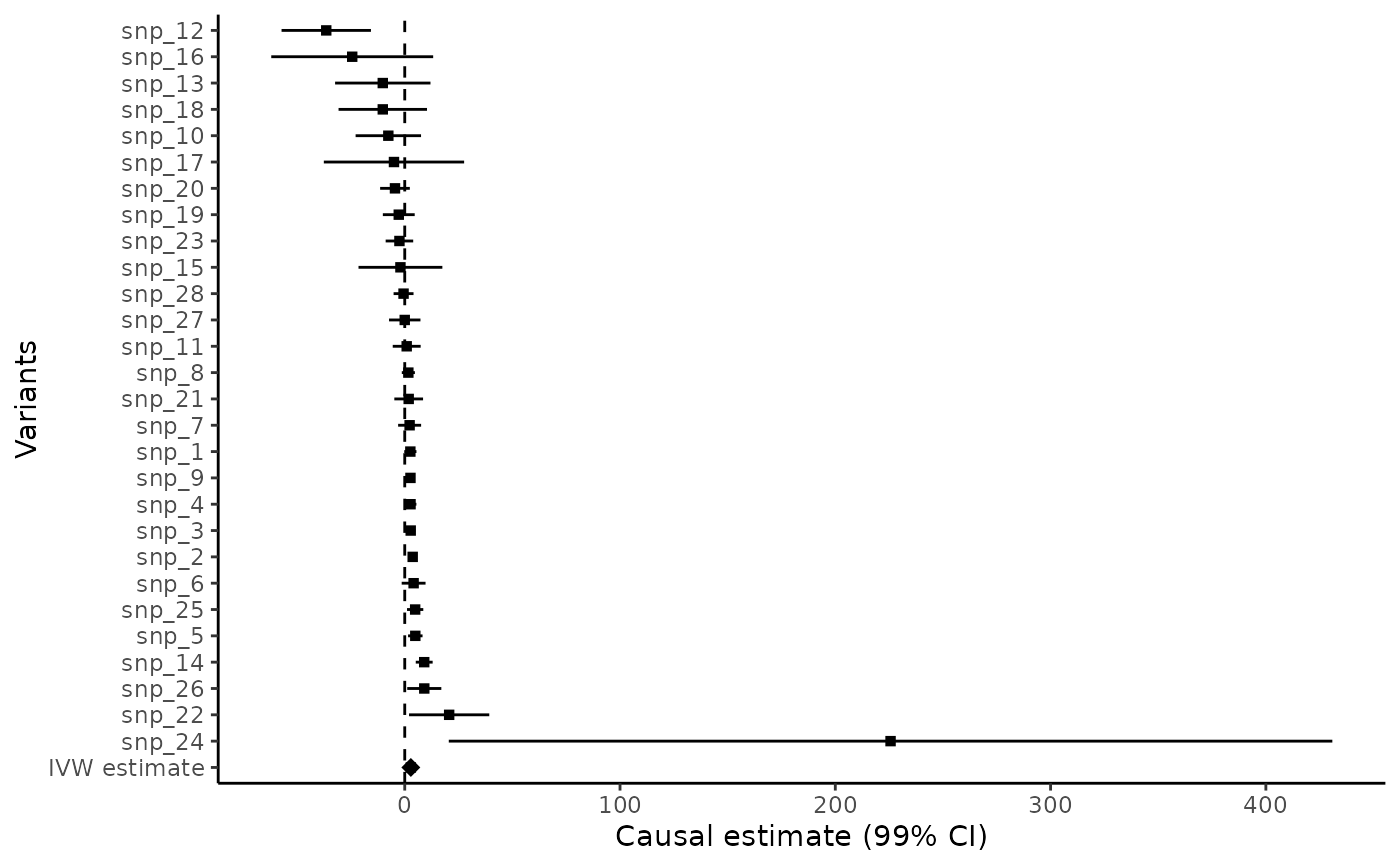 mr_forest(mr_input(bx = ldlc, bxse = ldlcse, by = chdlodds, byse = chdloddsse),
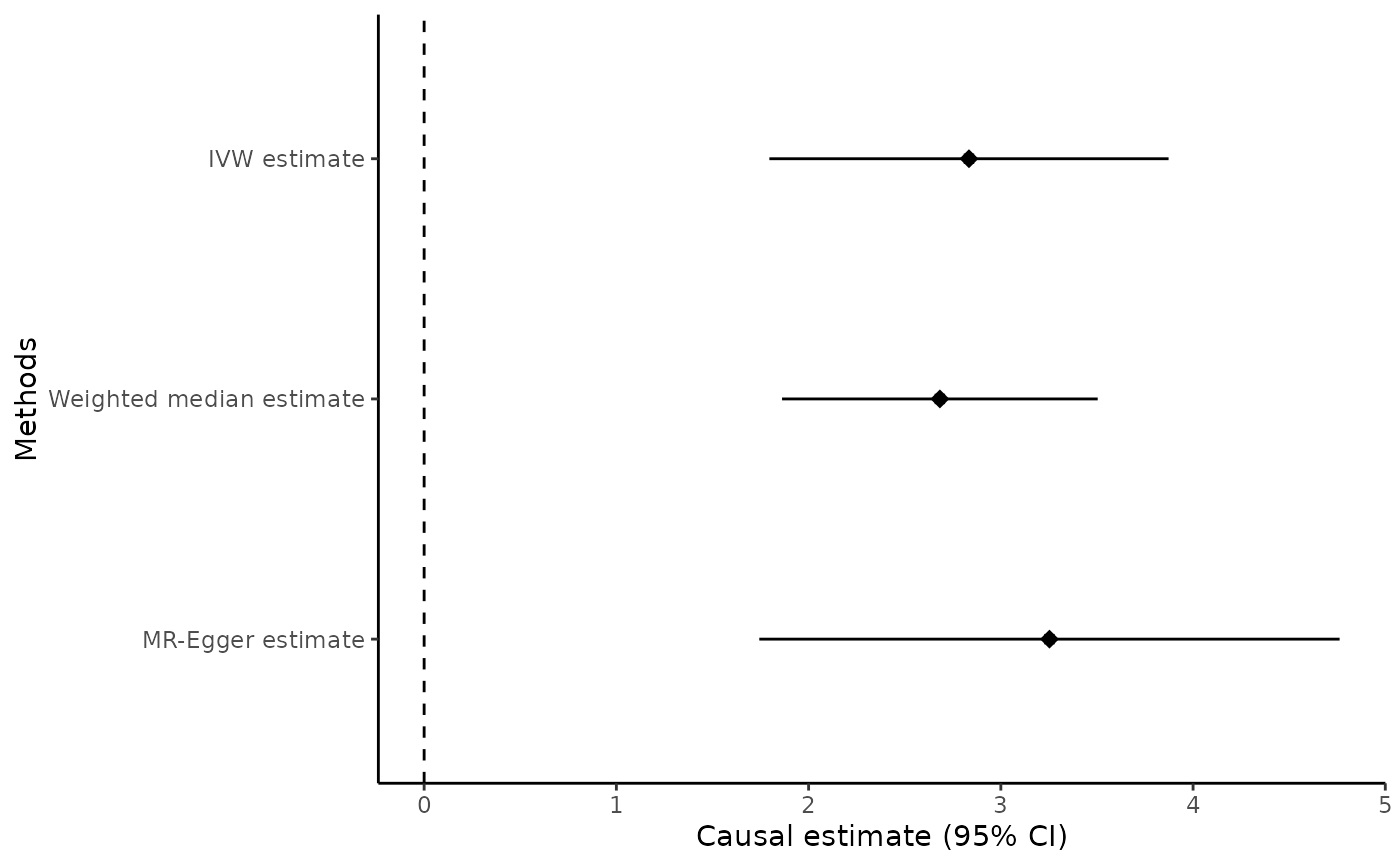
methods = c("ivw", "wmedian", "egger"), snp_estimates = FALSE)
mr_forest(mr_input(bx = ldlc, bxse = ldlcse, by = chdlodds, byse = chdloddsse),
methods = c("ivw", "wmedian", "egger"), snp_estimates = FALSE)
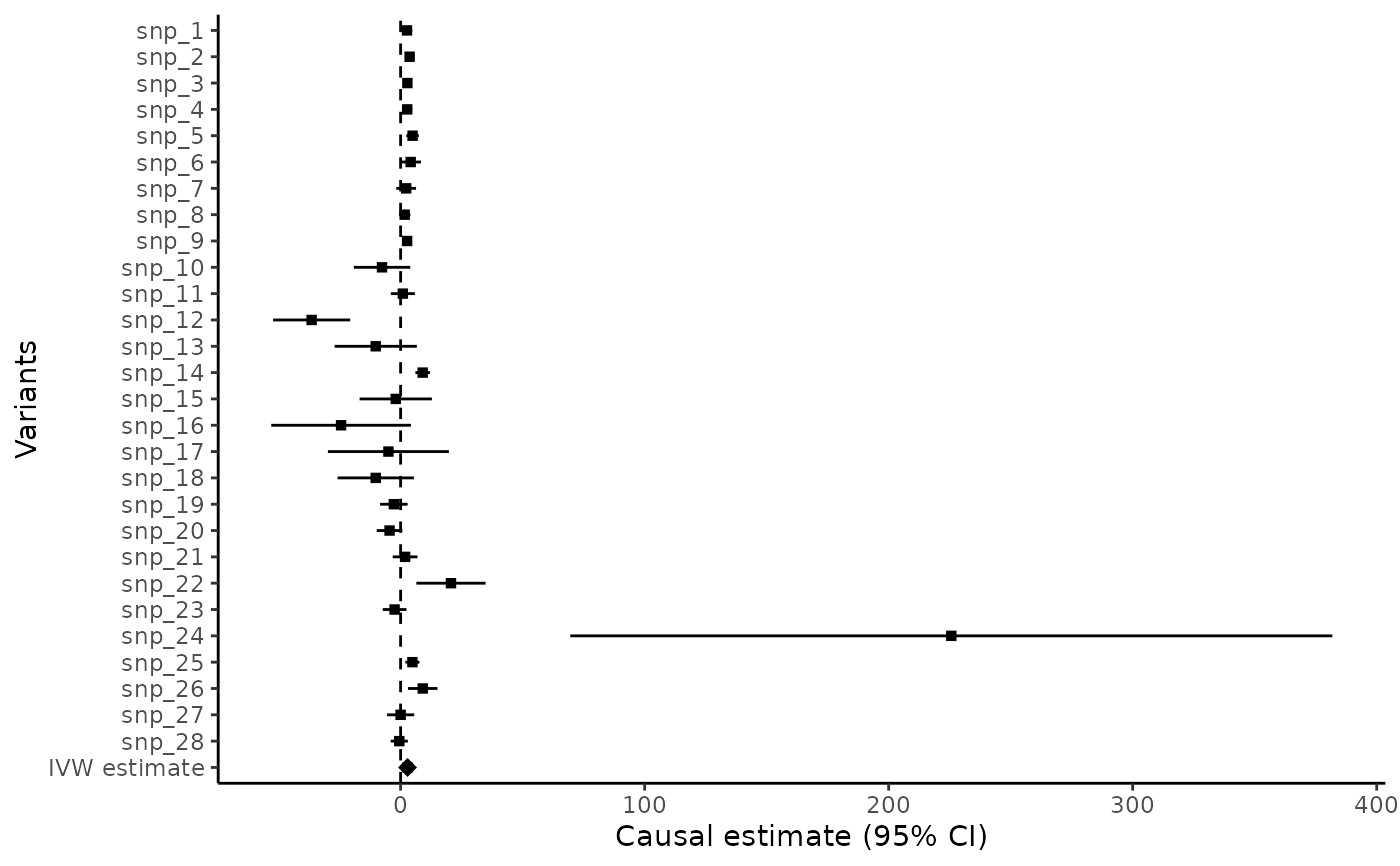
 forest = mr_forest(mr_input(bx = ldlc, bxse = ldlcse, by = chdlodds, byse = chdloddsse))
forest = mr_forest(mr_input(bx = ldlc, bxse = ldlcse, by = chdlodds, byse = chdloddsse))
 # how to change x-axis limits
# library(ggplot2)
# forest2 = forest + coord_cartesian(xlim=c(-5,5))
# forest2
# how to change x-axis limits
# library(ggplot2)
# forest2 = forest + coord_cartesian(xlim=c(-5,5))
# forest2